Low-rank GPs and the Predictive GP
Spatial Statistics - BIOSTAT 696/896
University of Michigan
Introduction
- We have introduced GP models
- Spatial GP models are priors over 2D surfaces (or 2D functions)
- Specifying the GP boils down to modeling covariance
- GPs can be used as part of a Bayesian hierarchical model
- We look for the posterior of a Bayesian model
- When the joint posterior is “difficult” we design MCMC algorithms to sample from it
- Models based on GPs scale poorly in terms of memory and flops and time
Scalable GP models
- We will now consider building special GPs that can scale to larger datasets
- There is no free lunch: scalability comes at a price
- As usual, we will need to consider trade-offs and make decisions based on our modeling goals
- Typically, we will restrict dependence across space, simplifying its structure
- By restricting spatial dependence we will speed up computations
- We will also lose in flexibility relative to an unrestricted GP
Graphical model interpretation of multivariate Gaussian random variables
Suppose we have a set of n random variables X_1, \dots, X_n
Call X = (X_1, \dots, X_n)^\top
Assume that X \sim N(0, \Sigma)
p(X_1, \dots, X_n) = p(X) = N(X; 0, \Sigma)
Graphical model interpretation of multivariate Gaussian random variables
- Using the properties of probability we can write \begin{align*} p(X_1, \dots, X_n) &= p(X_1) p(X_2 | X_1) \cdots p(X_n | X_1, \dots, X_{n-1})\\ &= \prod_i^n p(X_i | X_{[1:i-1]}) \end{align*} where X_{[1:i-1]} = (X_1, \dots, X_{i-1})^\top
- What is p(X_i | X_{[1:i-1]})?
Graphical model interpretation of multivariate Gaussian random variables
- What is p(X_i | X_{[i-1]})?
- p(X_i | X_{[1:i-1]}) is a conditional Gaussian distribution
- This is the same distribution that we get when we predict X_i using X_{[1:i-1]} N(X_i; H_i X_{[1:i-1]}, R_i) where H_i = \Sigma(i, [1:i-1]) \Sigma([1:i-1], [1:i-1])^{-1} and R_i = \Sigma(i, i) - H_i \Sigma([1:i-1], i).
Graphical model interpretation of multivariate Gaussian random variables
- Because p(X_1, \dots, X_n) = \prod_i^n p(X_i | X_{[i]}), we can draw a DAG according to which the joint density factorizes
- The number of edges cannot be any larger (for a DAG)
- If we added even just a single directed edge, the graphical model would form a cycle
- This DAG is dense
- We can attribute the computational complexity of GPs to the fact that their DAGs are dense
- Scaling GPs can be cast as a problem of sparsifying this DAG
- In other words, methods to remove edges from the DAG in a smart way
Graphical model interpretation of multivariate Gaussian random variables
- Draw a DAG for p(X_1, \dots, X_6) = p(X_1) p(X_2 |???) p(X_3 |???) \cdots p(X_6 | ???)
Graphical model interpretation of multivariate Gaussian random variables
- Draw a DAG for p(X_1, \dots, X_6)
- This DAG is dense
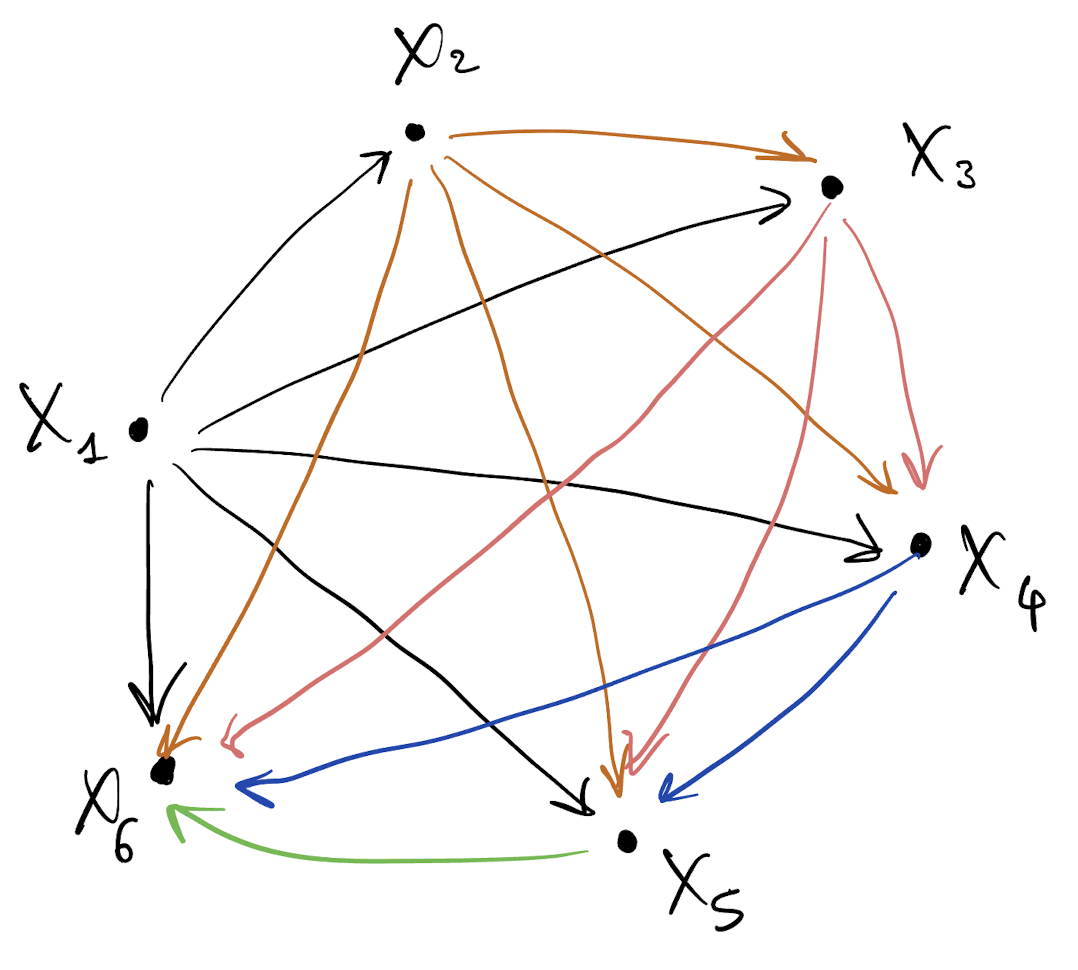
Graphical model interpretation of multivariate Gaussian random variables
- Draw a DAG for p(X_1, \dots, X_n)
- This DAG is dense
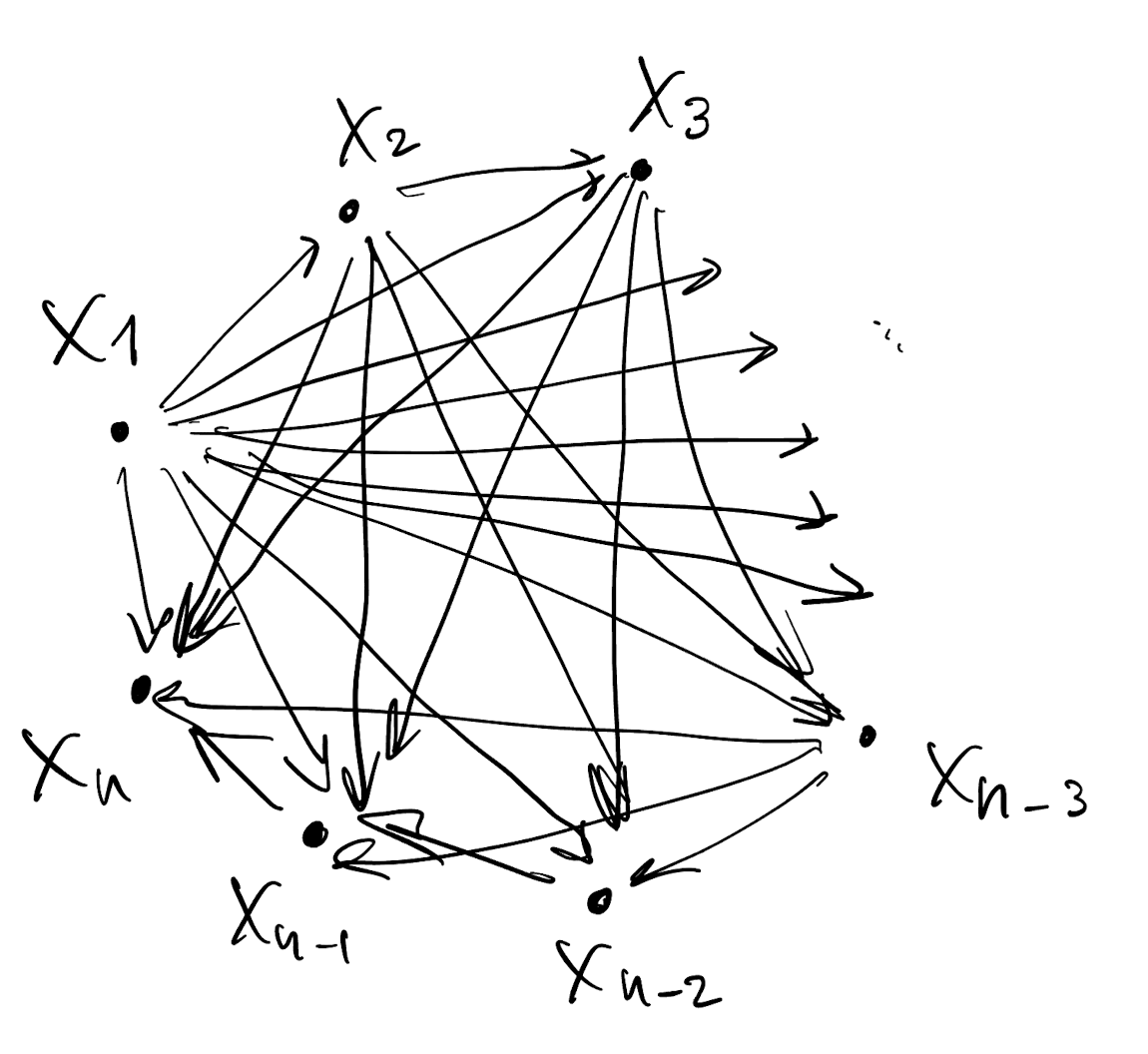
Graphical model interpretation of multivariate Gaussian random variables
- Draw a DAG for p(X_1, \dots, X_n)
- This DAG is dense
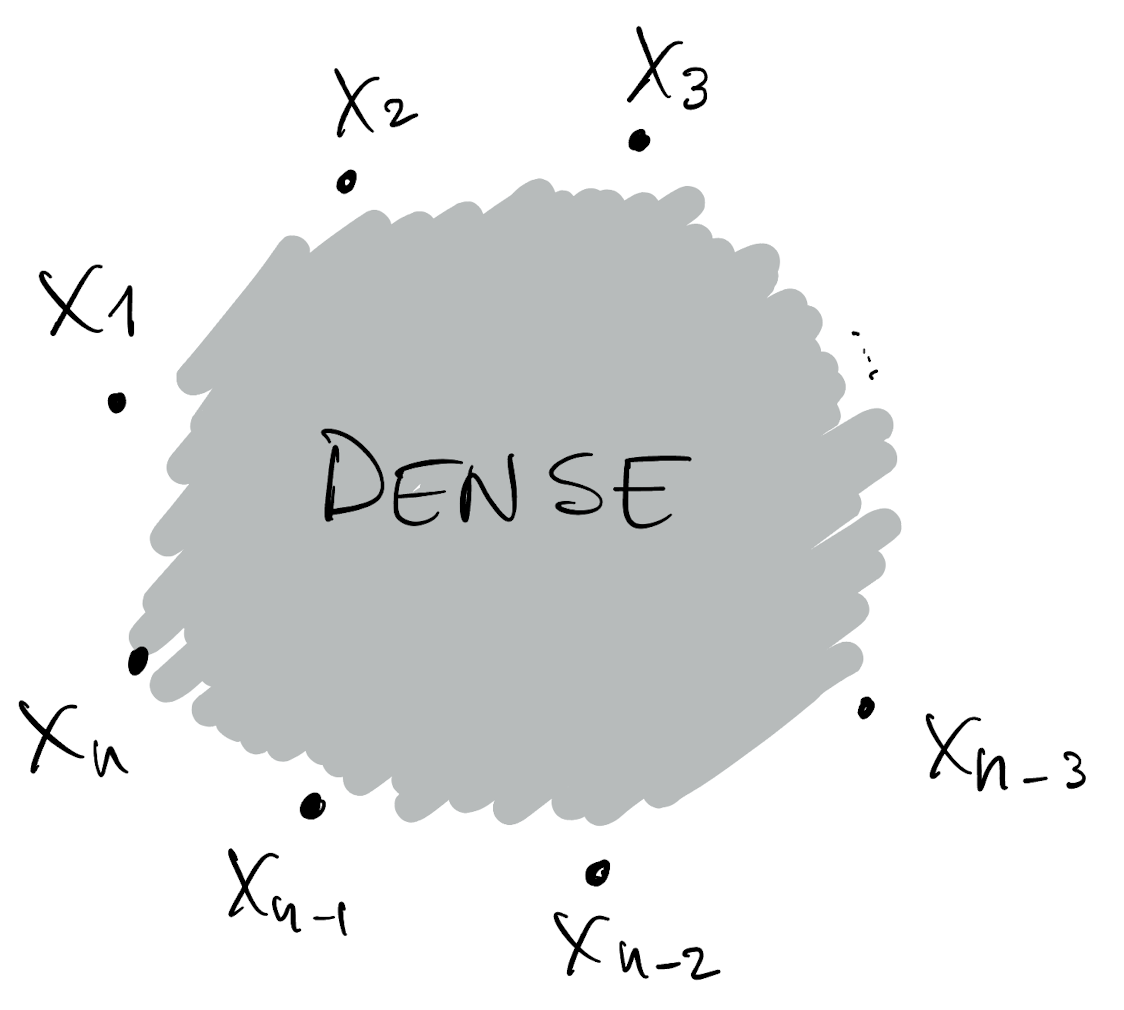
Low-rank Gaussian Processes
- Dense dependence follows from assuming a GP
- X(\cdot) \sim GP implies X_{\cal L} \sim N(0, \Sigma) implies dense DAG
- We want X(\cdot) \sim \text{???} leading to a simpler DAG that is sparse
Then, remember how we build a stochastic process:
- Define the joint distribution for any finite set of random variables
- Check Kolmogorov’s conditions
- Result: there is a stochastic process whose finite dimensional distribution is the one we chose
Low-rank Gaussian Processes
- Assume dense dependence for only a small set of variables
- All the others are assumed conditionally independent
- We assume the resulting joint density factorizes according to this DAG (see prelim slides)
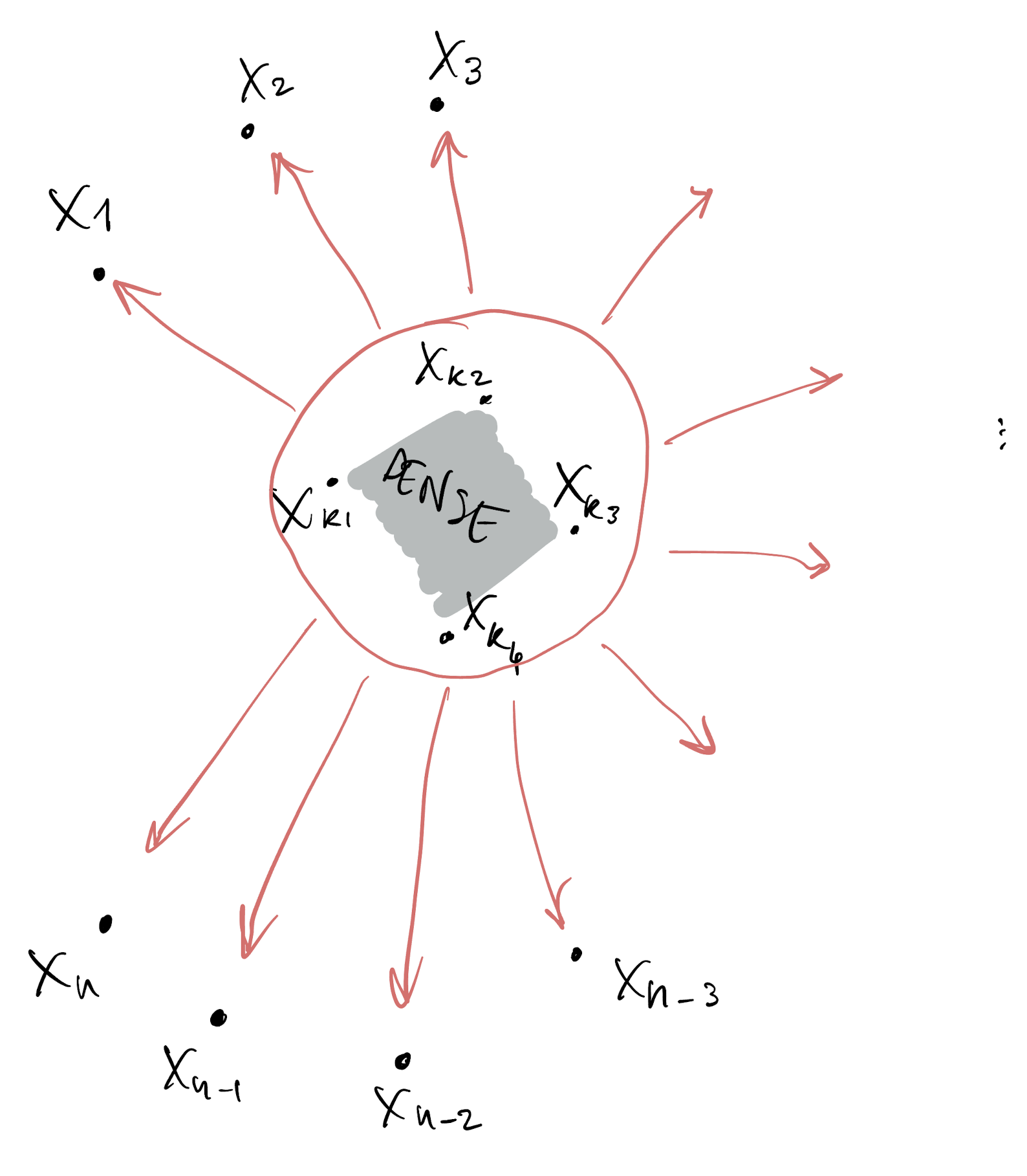
Low-rank Gaussian Processes
- Suppose from n variables we allow dense dependence for only k of them
- p(X_1, \dots, X_n) = p(X_1, \dots, X_k, X_{k+1}, \dots, X_n)
- The finite dimensional distribution we assume has the form
\begin{align*} p^*(X_1, \dots, X_n) &= p(X_1, \dots, X_k) \prod_{i=k+1}^n p(X_i | X_1, \dots, X_k) \end{align*}
- We then assume that all these densities are Gaussian
\begin{align*} p^*(X_1, \dots, X_n) &= N(X_{[1:k]}; 0, \Sigma_{[1:k]}) \prod_{i=k+1}^n N(X_i; H_i X_{[1:k]}, R_i) \end{align*}
where, as usual,
\begin{align*} H_i &= \Sigma_{i, [1:k]} \Sigma_{[1:k]}^{-1}\\ R_i &= \Sigma_i - H_i \Sigma_{i, [1:k]}^\top \end{align*}
Low-rank Gaussian Processes
- We allow dense dependence at a special set of locations which we call the knots
- We assume that the process at all other locations is conditionally independent on the value of the process at the knots
- We make a new joint density p^* that (unlike the original p density) factorizes according to the DAG we saw earlier
- To define a stochastic process, we need to define the finite dimensional distributions for any set \cal L of locations. Call \cal K the knots, assume \cal L \cap \cal K = \emptyset, then
\begin{align*} p^*(X_1, \dots, X_n) &= p^*(X_{\cal L}) \\ &= \int p(X_{\cal L} , X_{\cal K}) dX_{\cal K} \\ &= \int p(X_{\cal L} | X_{\cal K}) p(X_{\cal K}) dX_{\cal K} \\ &= \int \prod_{i \in \cal L} p(X_i | X_{\cal K}) p(X_{\cal K}) dX_{\cal K} \end{align*}
Low-rank Gaussian Processes
- We allow dense dependence at a special set of locations which we call the knots
- We assume that the process at all other locations is conditionally independent on the value of the process at the knots
- To define a stochastic process, we need to define the finite dimensional distributions for any set \cal L of locations. Call \cal K the knots, assume \cal K \subset \cal L and call \cal U = \cal L \setminus \cal K
\begin{align*} p^*(X_1, \dots, X_n) &= p^*(X_{\cal L}) = p(X_{\cal K}, X_{\cal U}) \\ &= p(X_{\cal U} | X_{\cal K}) p(X_{\cal K}) \end{align*}
Low-rank Gaussian Processes
From \begin{align*} p^*(X_1, \dots, X_n) &= \int \prod_{i \in \cal L} p(X_i | X_{\cal K}) p(X_{\cal K}) dX_{\cal K} \end{align*} we assume all densities are Gaussian
\begin{align*} p^*(X_1, \dots, X_n) &= \int \prod_{i \in \cal L} N(X_i; H_i X_{\cal K}, R_i ) N(X_{\cal K}; 0, C_{\cal K}) dX_{\cal K} \\ &= \int N(X_{\cal L}; H_{\cal L} X_{\cal K}, D_{\cal L} ) N(X_{\cal K}; 0, C_{\cal K}) dX_{\cal K} \end{align*}
- p^*(X_1, \dots, X_n) = ????
- We define D_{\cal L} as the diagonal matrix collecting all R_i.
Low-rank Gaussian Processes
\begin{align*} p^*(X_1, \dots, X_n) &= \int N(X_{\cal L}; H_{\cal L} X_{\cal K}, D_{\cal L} ) N(X_{\cal K}; 0, C_{\cal K}) dX_{\cal K} \end{align*} We can write this as
\begin{align*} p(X_{\cal K}): \qquad X_{\cal K} &\sim N(0, C_{\cal K})\\ p(X_{\cal L} | X_{\cal K}): \qquad X_{\cal L} &= H_{\cal L} X_{\cal K} + \eta_{\cal L}, \qquad \eta_{\cal L} \sim N(0, D_{\cal L}) \end{align*} Therefore \begin{align*} p(X_{\cal L}): \qquad X_{\cal L} &\sim N(0, H_{\cal L} C_{\cal K} H_{\cal L}^\top + D_{\cal L}) \end{align*}
And here we see that
H_{\cal L} C_{\cal K} H_{\cal L}^\top = C_{\cal L, \cal K} C_{\cal K}^{-1} C_{\cal K} C_{\cal K}^{-1} C_{\cal K, \cal L} = C_{\cal L, \cal K} C_{\cal K}^{-1} C_{\cal K, \cal L}
Low-rank Gaussian Processes
From the previous slide: if X(s) and X(s') are both realizations of a low-rank GP with knots \cal K, then if s\ne s' Cov(X(s), X(s')) = C(s, \cal K) C_{\cal K}^{-1} C(\cal K, s') \ne C(s, s')
Where C(s, \cal K) is our chosen covariance function. Whereas if s=s' then Cov(X(s), X(s')) = C(s, \cal K) C_{\cal K}^{-1} C(\cal K, s') + D_{s} = C(s, s)
Notice anything? What is Cov(X(s), X(s')) in the unrestricted GP?
Low-rank Gaussian Processes: induced covariance function
If X(\cdot) is a low-rank GP constructed using covariance function C(\cdot, \cdot), then
- Cov(X(s), X(s')) \neq C(s, s')
- Equivalently, X(\cdot) is an unrestricted GP using the covariance function \tilde{C}(\cdot, \cdot) where
\tilde{C}(s, s') = Cov(X(s), X(s')) = C(s, \cal K) C_{\cal K}^{-1} C(\cal K, s') \qquad \text{if } s\neq s' \tilde{C}(s, s') = C(s, s') \qquad \text{if } s= s'
In the literature
What we have seen in the previous slides corresponds to Finley, Sang, Banerjee, Gelfand (2009) doi:10.1016/j.csda.2008.09.008.
- Quiñonero-Candela and Rasmussen (2005)
https://www.jmlr.org/papers/v6/quinonero-candela05a.html - Banerjee, Gelfand, Finley and Sang (2008)
doi:10.1111/j.1467-9868.2008.00663.x - The ML literature typically refers to these models as sparse GPs
- “Knots” are sometimes called “sensors” or “inducing points”
In a regression model
- Assume observed locations are \cal L
- Make all the usual assumptions for regression and prior
- The only change: w \sim LRGP with covariance function C(\cdot, \cdot) and knots \cal K:
Y = X\beta + w + \varepsilon We can write the model as
Y = X\beta + H w_{\cal K} + \eta + \varepsilon where H = C_{\cal L, \cal K} C_{\cal K}^{-1} and \eta \sim ???
- If we omit \eta from the model, then we are fitting a regression model based on the Predictive Gaussian Process as introduced in Banerjee, Gelfand, Finley and Sang (2008)
- If we include \eta then we are fitting a Modified Predictive Gaussian Process model as in Finley, Sang, Banerjee, Gelfand (2009). Equivalently,
Y = X\beta + H w_{\cal K} + \varepsilon^* \qquad \text{where} \qquad \varepsilon^* \sim N(0, \tau^2 I_n + D_{\cal L})
Low-rank Gaussian Processes in R
- In R, the
spBayespackage implements the (modified) predictive process as we have seen here - Use argument
knots=...inspLM - Everything we said earlier about
spLMremains true even when using the predictive process